Irrespective of the intention and final goal (either taxonomy, biology, or applications), every Trichoderma diversity research starts from the identification of sampled species, i.e., use of the existing taxonomy.
Below, are a few practical recommendations that aim to provide answers to the most frequent questions that were addressed to ICTT in our practice of molecular identification of Trichoderma and also aid in the evaluation of Trichoderma diversity studies by reviewers, editors, and decision-makers in organizations financing such studies. The first version of these recommendations was published in Cai and Druzhinina (2021).
Key references on fungal taxonomy
Reading of the following literature is highly recommended before approaching Trichoderma taxonomy:
The latest edition of Chapter F by (May et al. 2019) in the Code, (https://www.iapt-taxon.org/nomen/pages/main/chapter_f.html) and the Code (https://www.iapt-taxon.org/nomen/main.php). Additionally, become familiar with the original requirements regarding the deposition of reference materials and types in public databases, naming, and imaging. It is also recommended to address the most recent fungal taxonomy and fungal DNA Barcoding guidelines (Lucking et al., 2021; Lücking et al. 2020; Schoch et al. 2020; Vu et al. 2019; May et al. 2019) and the original publication on the new species description standard in fungi by Seifert and Rossman (2010). Independent of the publication date, taxonomic descriptions of all related species, taxonomic revisions of the related infrageneric groups, and non-taxonomic literature on the species that belong to the group of interest should be investigated.
Consulting with the experienced experts
Specialists in fungal taxonomy and nomenclature can be contacted through the International Committee of Taxonomy of Fungi (www.fungaltaxonomy.info), the Nomenclature Committee for Fungi (NCF) (https://www.ima-mycology.org/nomenclature/nomenclature-committee-fungi), the International Mycological Association (IMA) (https://www.ima-mycology.org/), or through the regional Member Mycological Organizations http://www.ima-mycology.org/society/member-mycological-organizations or also listed in Wikipedia (https://en.wikipedia.org/wiki/Category:Mycology_organizations).
Experts on Trichoderma taxonomy can be contacted through the contact form on this website.
Trichodermadiversity surveys and DNA Barcoding
(1) Do not expect high diversity of Trichoderma in soil. It is not a soil fungus (Friedl and Druzhinina 2012; Kubicek et al. 2019).
(2) Do not add fungicides to the isolation medium. The growth of numerous rare species is reduced by such fungicides as Rose Bengal and others (I.S. Druzhinina, unpublished).
(3) Do not rely on phenotypical or morphological similarity for grouping the strains for DNA Barcoding. Many Trichoderma spp. are morphologically identical (cryptic) (Jaklitsch 2009, 2011; Jaklitsch and Voglmayr 2015; Chaverri et al. 2015).
(4) Do not rely on ITS for the preliminary grouping of isolates for the subsequent DNA Barcoding. Many sister species share the same ITS phylotype (Druzhinina et al. 2012; Sandoval-Denis et al. 2014; Druzhinina et al. 2005). The probability to isolate two or more of such species from the same habitat is considerable because several related Trichoderma species co-occur (Komoń-Zelazowska et al. 2007; Friedl and Druzhinina 2012) and therefore cannot be distinguished by ITS.
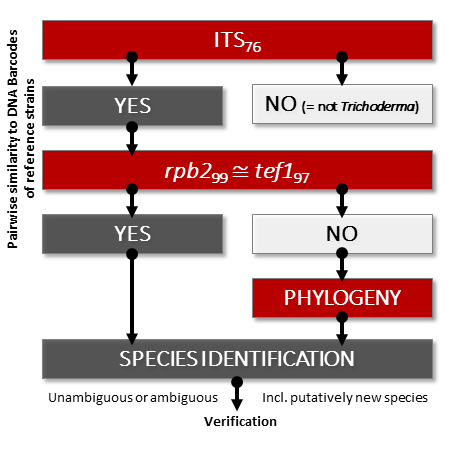
(5) Sequence of DNA barcoding fragments of ITS, tef1, and rpb2 for all isolates. Consider selecting primer pairs of tef1 that will guarantee the sequencing of the diagnostic region (see examples here, note other primer pairs listed in Rahimi et al., 2020).
(6) Use on-line tools and public databases for the preliminary analysis of the obtained DNA barcodes [such as MIST, (Dou et al. 2020) or NCBI BLAST, (Ye et al. 2006)]. These analyses will help to reveal genetically unique or common isolates. Consider the results that were obtained using automated tools as preliminary or putative molecular identification. CURRENTLY, THERE ARE NO ONLINE TOOLS THAT CAN PROVIDE A RELIABLE IDENTIFICATION OF TRICHODERMA SPECIES.
(7) Follow the molecular identification protocol for a single Trichoderma isolate including the validation step.
(8) Use original taxonomic literature and the metadata for the query strains (morphology, physiology, ecology, biogeography, occurrence) for the biological verification of the identification results. Assign ambiguous identification if the biological verification fails.
(9) While depositing sequences in public databases, taxonomic accuracy is more appreciated over precision. For ambiguous results T. sp. [strain ID] is preferred over the assignment of an ambiguously identified species name. Alternatively, use T. aff. [closest species] or T. cf. [one of several close species] strain ID formats.
(10) If the molecular identification and subsequent biological verification suggest that a putative new species has been detected, consider the following requirements:
– Check the compliance with the Code.
– Verify Latin grammar for the new species name.
– Consider intraspecific polymorphism (more than one strain or specimen).
– Apply GCPSR concept (compulsory consideration of single locus trees).
– Aim to use the polyphasic approach that implies detailed comparative ecophysiological characterization of the putative new species and closely related taxa.
– Deposit the maximum number of DNA barcodes for each isolate and for more than a single isolate. Collect and provide the most explicit metadata.
– Test the identifiability of the strain.
– In ambiguous case, consult with experts.
(11) Obtain the most precise species identification before subjecting a Trichoderma strain for a WGS. Genomics is highly useful for the study of fungal biology, but its applicability in taxonomy and identification is still limited.
(12) Verify the use of Trichoderma gene nomenclature